Please see Chapter 11 Biotechnology Principles And Processes Exam Questions Class 12 Biology below. These important questions with solutions have been prepared based on the latest examination guidelines and syllabus issued by CBSE, NCERT, and KVS. We have provided Class 12 Biology Questions and answers for all chapters in your NCERT Book for Class 12 Biology. These solved problems for Biotechnology Principles And Processes in Class 12 Biology will help you to score more marks in upcoming examinations.
Exam Questions Chapter 11 Biotechnology Principles And Processes Class 12 Biology
Very Short Answer Questions
Question. Name the host cells in which micro-injection technique is used to introduce an alien DNA.
Ans. Animal cells.
Question. Do eukaryotic cells have restriction endonucleases? Justify your answer.
Ans. Eukaryotic cells have no restriction enzymes as the DNA molecules of eukaryotes are heavily methylated. It is present in prokaryotic cell (like bacteria) where these act as defence mechanism to restrict the growth of bacteriophages.
Question. Write the name of the enzymes that are used for isolation of DNA from bacterial and fungal cells respectively for Recombinant DNA technology.
Ans. Bacterial cell is treated with enzyme lysozyme. Fungal cell is treated with chitinase.
Question. What are palindromes?
Ans. Palindromes are group of letters (sequences) that read same both in forward and backward direction.
Question. What is the cell that receives a recombinant gene called?
Ans. Competent host cell/recipient cell.
Question. What is recombinant DNA?
Ans. Recombinant DNA is the DNA formed by combining DNAs from two different organisms.
Question. Identify the reason for selection of DNA polymerase from Thermus aquaticus for Polymerase Chain Reaction.
Ans. DNA polymerase from Thermus aquaticus remains active during the high temperature induced denaturation of double stranded DNA.
Question. What would be the molar concentration of human DNA in a human cell? Consult your teacher.
Ans. The average molecular weight of a nucleotide pair is 330 dalton. Genome size of a diploid human cell is around 6 × 109 bp. The concentration of DNA will be 330 × 6 × 109g/mol.
The molarity can then be calculated as: Sample DNA concentration (in g) /330×6×10 g/mol.
Short Answer Questions
Question. Explain the work carried out by Cohen and Boyer that contributed immensely in biotechnology.
Ans. Stanley Cohen and Herbert Boyer in 1972 constructed the first recombinant DNA. They isolated the antibiotic resistance gene by cutting out a piece of DNA from the plasmid of a bacterium which was responsible for conferring antibiotic resistance. The cut piece of DNA was then linked with the plasmid DNA of Salmonella typhimurium and transferred to E. coli for transformation.
Question. Can you list 10 recombinant proteins which are used in medical practice? Find out where they are used as therapeutics (use the internet).
Ans.
| S.No. | Recombinant proteins | Therapeutic uses |
| (i) | Human Insulin (Humulin) | Treatment of diabetes type 1. |
| (ii) | Tissue Plasminogen Activator | Treatment for acute myocardial infarction; dissolves blood clot after heart attack and stroke. |
| (iii) | DNase | Treatment of cystic fibrosis. |
| (iv) | Platelet Growth Factor | Stimulation of wound healing. |
| (v) | Calcitonin | Treatment of rickets. |
| (vi) | Reo Pro | Prevention of blood clots. |
| (vii) | Hirudin | Used as an anticoagulant. |
| (viii) | Interferon (α, b and γ) | Treatment of viral infection and cancer. |
| (ix) | Chorionic Gonadotropin | Treatment of infertility. |
| (x) | Interleukins | Enhancing activity of immune system. |
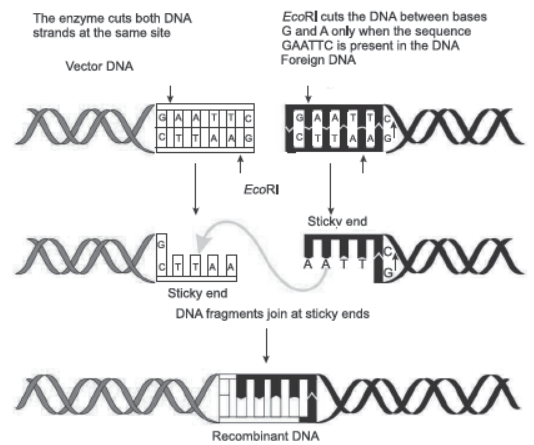
Question. Why are molecular scissors so called? Write their use in biotechnology.
Ans. (a) The restriction endonucleases are called molecular scissors, as they cut the DNA segments at
particular locations, e.g., EcoRI.
(b) The restriction enzymes cut the DNA strands a little away from the centre of the palindromic sites, but between the same two bases on the opposite strands. This leaves single stranded portions with overhanging stretches called sticky ends on each strand as they form hydrogen bonds with their complementary cut counterparts. This stickiness at the ends facilitates the action of the enzyme DNA ligase.
Question. Explain the role of the enzyme EcoRI in recombinant DNA technology.
Ans. EcoRI inspects length of DNA and recognises specific palindromic nucleotide sequences. It then binds with DNA and cuts each of the two strands of double helix at specific points.
Refer to Basic Concepts Point 4 (Mechanism of Action of Endonucleases).
The first endonuclease discovered was HindII.
Question. Write the convention used for naming restriction enzymes.
OR
Explain with the help of a suitable example the naming of a restriction endonuclease.
Ans. The convention for naming restriction enzymes is that the first letter to the name comes from the Genus and the second two letters come from species and third letter indicates the strain of the prokaryotic cell from which they are isolated e.g., EcoRI comes from Escherichia coli RI, here R stands for the strain and I follows the order in which the enzyme was isolated.
Question. Make a chart (with diagrammatic representation) showing a restriction enzyme, the substrate
DNA on which it acts, the site at which it cuts DNA and the product it produces.
Ans.
Question. Name the natural source of agarose. Mention one role of agarose in biotechnology.
Ans. The natural source of agarose is sea weed. Agarose is a natural polymer. It is used to develop the matrix for gel electrophoresis. It helps in the separation of DNA fragments based on their size.
Question. What are ‘cloning sites’ in a cloning vector? Explain their role. Name any two such sites in pBR322.
Ans. Cloning sites are the recognition sites on plasmid. The restriction enzymes recognise these sites for cutting and ligation of alien DNA at this place. For example,E coRI, BamHI.
Question. (a) Mention the difference in the mode of action of exonuclease and endonuclease.
(b) How does restriction endonuclease function?
Ans. (a) Exonuclease removes nucleotides from the ends of DNA whereas endonuclease cuts at specific positions within DNA at specific positions.
(b) Restriction endonuclease recognises and cuts specific palindromic nucleotide sequences in the DNA.
Question. Why and how bacteria can be made ‘competent’?
Ans. Bacteria are made competent to accept the DNA or plasmid molecules. This is done by treating them with specific concentration of a divalent cation such as calcium to increase pore size in cell wall. The cells are then incubated with recombinant DNA on ice, followed by placing them briefly at 42°C and then putting it back on ice.
Question. How does a restriction nuclease function? Explain.
Ans. Restriction nuclease cuts DNA at specific sites. Nucleases are of two types exonuclease and endonuclease.
Exonuclease cuts DNA at the ends, whereas endonuclease cuts at specific sites within DNA.
Question. You have created a recombinant DNA molecule by ligating a gene to a plasmid vector. By mistake, your friend adds exonuclease enzyme to the tube containing the recombinant DNA.
How will your experiment get affected as you plan to go for transformation now?
Ans. The experiment will not likely be affected as recombinant DNA molecule is circular and closed, with no free ends. Hence, it will not be a substrate for exonuclease enzyme which removes nucleotides from the free ends of DNA.
Question. A plasmid without a selectable marker was chosen as vector for cloning a gene.
Ans. In a gene cloning experiment, first a recombinant DNA molecule is constructed, where the gene of interest is ligated to the vector and introduced inside the host cell (transformation). Since, not all the cells get transformed with the recombinant/plasmid DNA, in the absence of selectable marker, it will be difficult to distinguish between transformants and non-transformants, because role of selectable marker is in the selection of transformants.
Question. Explain palindromic nucleotide sequence with the help of a suitable example.
Ans. The palindrome in DNA is a sequence of base pairs that reads same on the two strands when orientation of reading is kept the same. For example, the following sequences reads the same on the two strands in 5′ 3′ direction. This is also true if it is read in the 3′ 5′ direction.
5′ — — GAATTC — — 3′
3′ — — CTTAAG — — 5′
Long Answer Questions
Question. (a) Draw the figure of vector pBR322 and label the following:
(i) Origin of replication
(ii) Ampicillin resistance site
(iii) Tetracycline resistance site
(iv) BamH1 restriction site
(b) Identify the significance of origin of replication.
Ans. (a) Refer to
(b) Origin of replication is responsible for controlling the copy number of the DNA sequence inserted.
Question. Many copies of a specific gene of interest are required to study the detailed sequencing of bases in it. Name and explain the process that can help in developing large number of copies of this gene of interest.
Ans. Polymerase Chain reaction (PCR).
Amplification of gene of interest using PCR
• The Polymerase Chain Reaction (PCR) is a reaction in which amplification of specific DNA sequences is carried out in vitro.
• This technique was developed by Kary Mullis in 1985, and for this, he received Nobel Prize for Chemistry in 1993.
Question. (a) In pBR322, foreign DNA has to be introduced in tetR region. From the restriction enzymes given below, which one should be used and why?
PvuI, EcoRI, BamHI
(b) Give reasons, why the other two enzymes cannot be used.
Ans. (a) BamHI should be used, as restriction site for this enzyme is present in tetR region.
(b) PvuI will not be used, as restriction site for this enzyme is present in ampR region (not in tetR).
EcoRI will not be used, as restriction site for this enzyme is not present in selectable marker tetR.
Question. Name and explain the technique used for separating DNA fragments and making them available for biotechnology experiments.
OR
How are the DNA fragments separated and isolated for DNA fingerprinting? Explain.
Ans. QU Gel electrophoresis is a technique for separating DNA fragments based on their size.
• Firstly, the sample DNA is cut into fragments by restriction endonucleases.
• The DNA fragments are seen as orange coloured bands.
• The separated bands of DNA are cut out and extracted from the gel piece. This step is called elution.
• The purified DNA fragments are used to form recombinant DNA which can be joined with cloning vectors.
Question. How does b-galactosidase coding sequence act as a selectable marker? Explain. Why is it a preferred selectable marker to antibiotic resistance genes?
Ans. When a recombinant DNA is inserted within the coding sequence of the enzyme b-galactosidase, it results into inactivation of the enzyme. The presence of a chromogenic substrate gives blue coloured colonies if the plasmid in the bacteria does not have an insert, whereas presence of insert do not produce any colour.
Selection of recombinants due to inactivation of antibiotics is a cumbersome procedure because it requires simultaneous plating on two plates having different antibiotics. Therefore, selectable markers are preferred for selection of recombinants.
Question. Explain the importance of (a) ori, (b) ampR and (c) rop in the E. coli vector shown below:
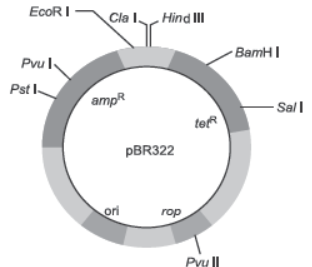
Ans. (a) ori: Ori is a sequence from where replication starts and any piece of DNA when linked to this sequence can be made to replicate within the host cells. It is also responsible for controlling the copy number of the linked DNA.
(b) ampR: The ligation of alien DNA is carried out at a restriction site present in any antibiotic resistance gene.
(c) rop: It codes for the proteins involved in the replication of the plasmid.
Question.
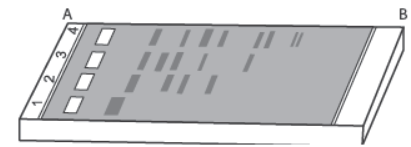
(a) Mark the positive and negative terminals.
(b) What is the charge carried by DNA molecule and how does it help in its separation?
(c) How the separated DNA fragments are finally isolated?
Ans. (a) Positive terminal – ‘B’
Negative terminal – ‘A’
(b) DNA is negatively charged. Because of its negative charge, DNA moves towards the positive electrode (anode).
(c) The separated DNA fragments are separated by elution. The separated bands of DNA are cut out from the agarose gel and extracted from the gel piece.
Question. List the key tools used in recombinant DNA technology.
Ans. The key tools used in recombinant DNA technology are:
(i) Restriction enzym e s (ii) Polymerase enzyme
(iii) Ligase enzyme (iv) Vectors
(v) Host organism/cell.
Question. A plasmid DNA and a linear DNA (both of the same size) have one site for a restriction endonuclease. When cut and separated on agarose gel electrophoresis, plasmid shows one DNA band while linear DNA shows two fragments. Explain.
Ans. It is because plasmid is a circular DNA molecule. When cut with enzyme, it becomes linear but does not get fragmented. Whereas, a linear DNA molecule gets cut into two fragments. Hence, a single DNA band is observed for plasmid while two DNA bands are observed for linear DNA in agarose gel. (Img 420)
Question. A mixture of fragmented DNA was electrophoresed in agarose gel. After staining the gel with ethidium bromide, no DNA bands were observed. What could be the reason?
Ans. The reasons that could be possible are as follows:
(i) DNA sample that was loaded on the gel may have got contaminated with nuclease (exo- or endo- or both) and completely degraded.
(ii) Electrodes were put in opposite orientation in the gel assembly, i.e., anode towards the wells (where DNA sample is loaded). Since DNA molecules are negatively charged, they move towards anode and hence move out of the gel instead of moving into the matrix of gel.
(iii) Ethidium bromide was not added at all or was not added in sufficient concentration and so DNA was not visible.
(iv) After staining with Ethidium bromide it was not observed under UV.

